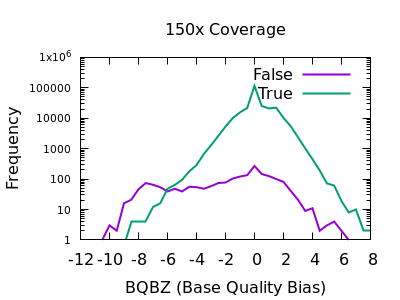
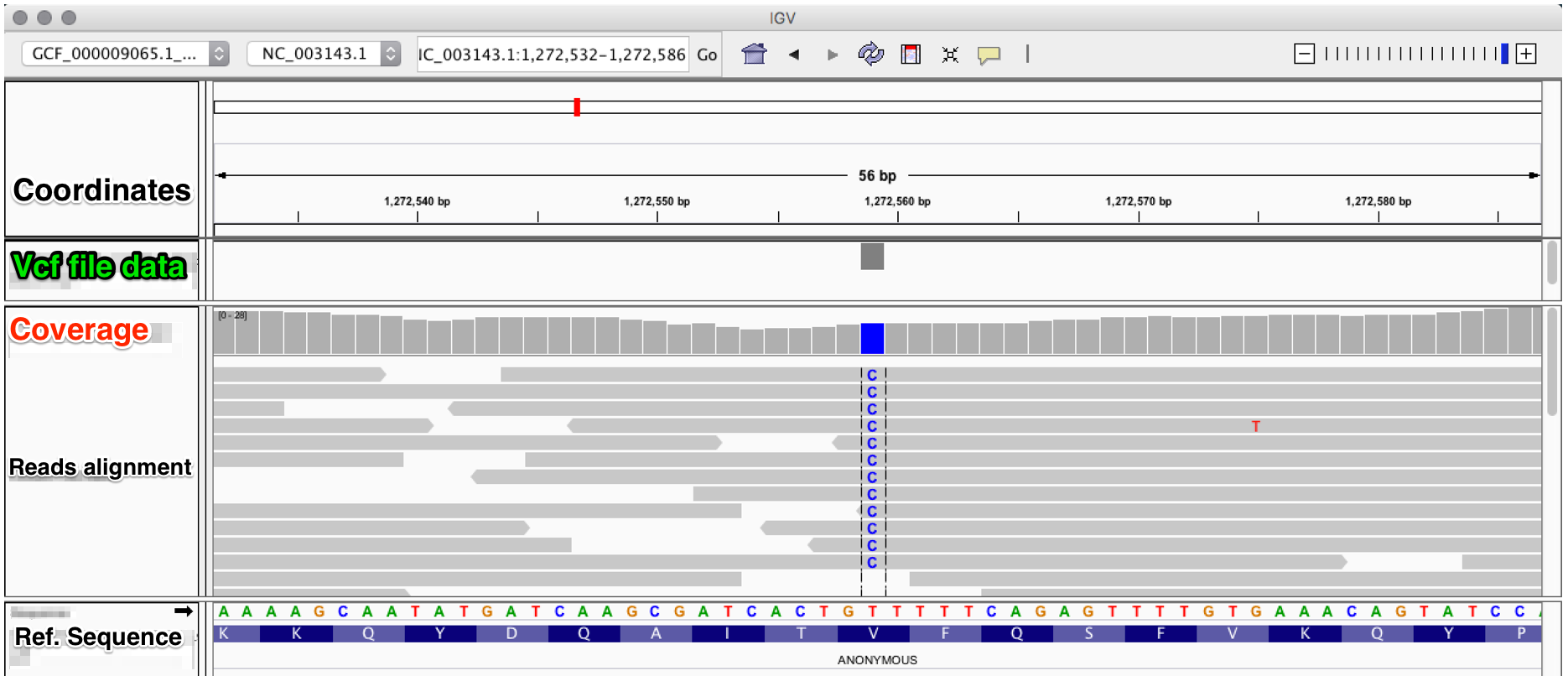
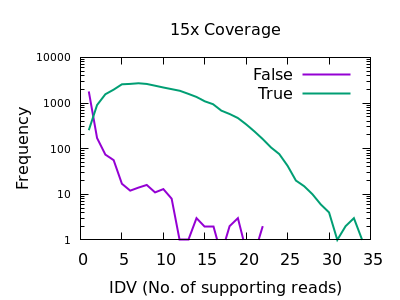
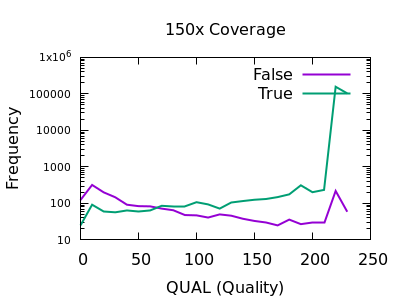
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports
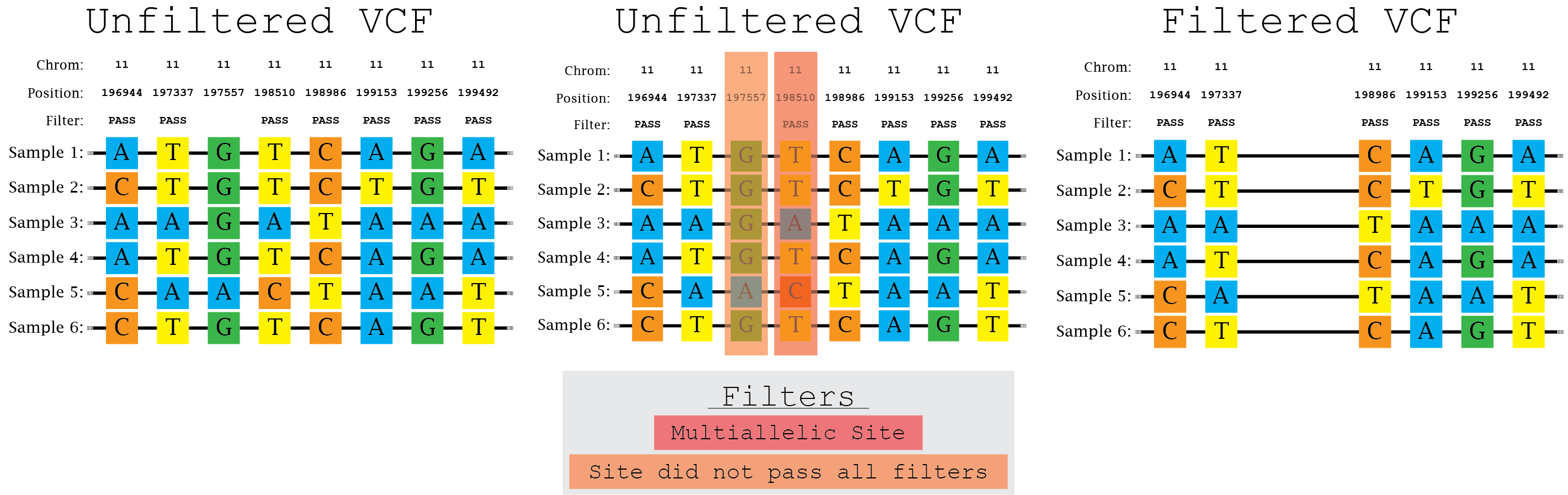
a) Filtering different variant callers VCF output for SARS-CoV-2 data.... | Download Scientific Diagram
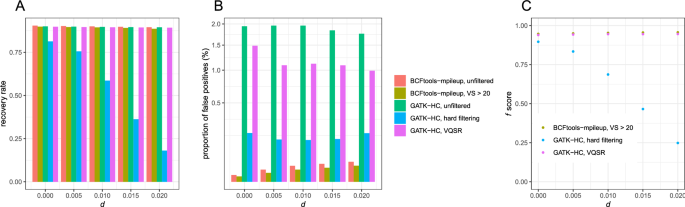
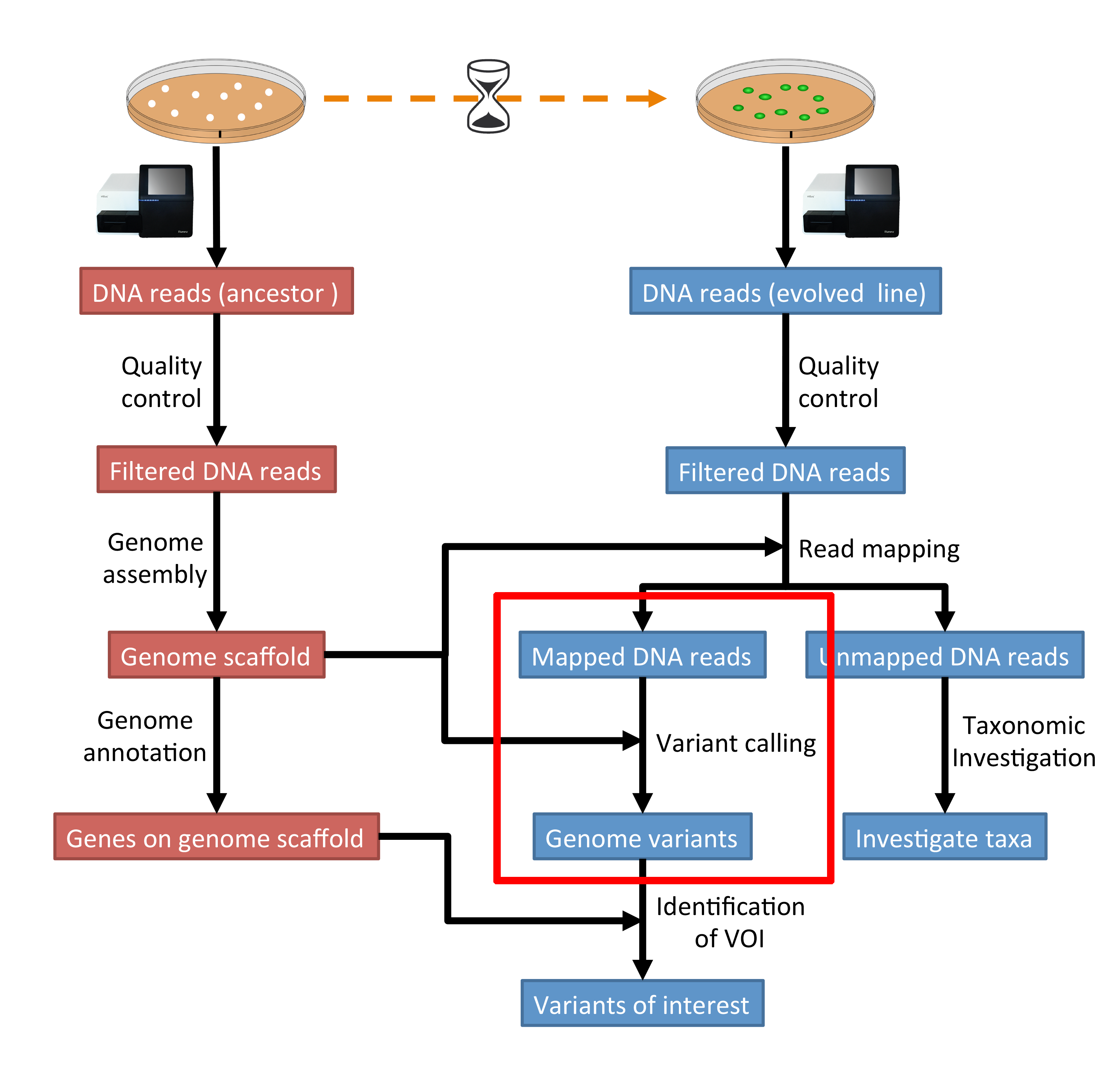
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports

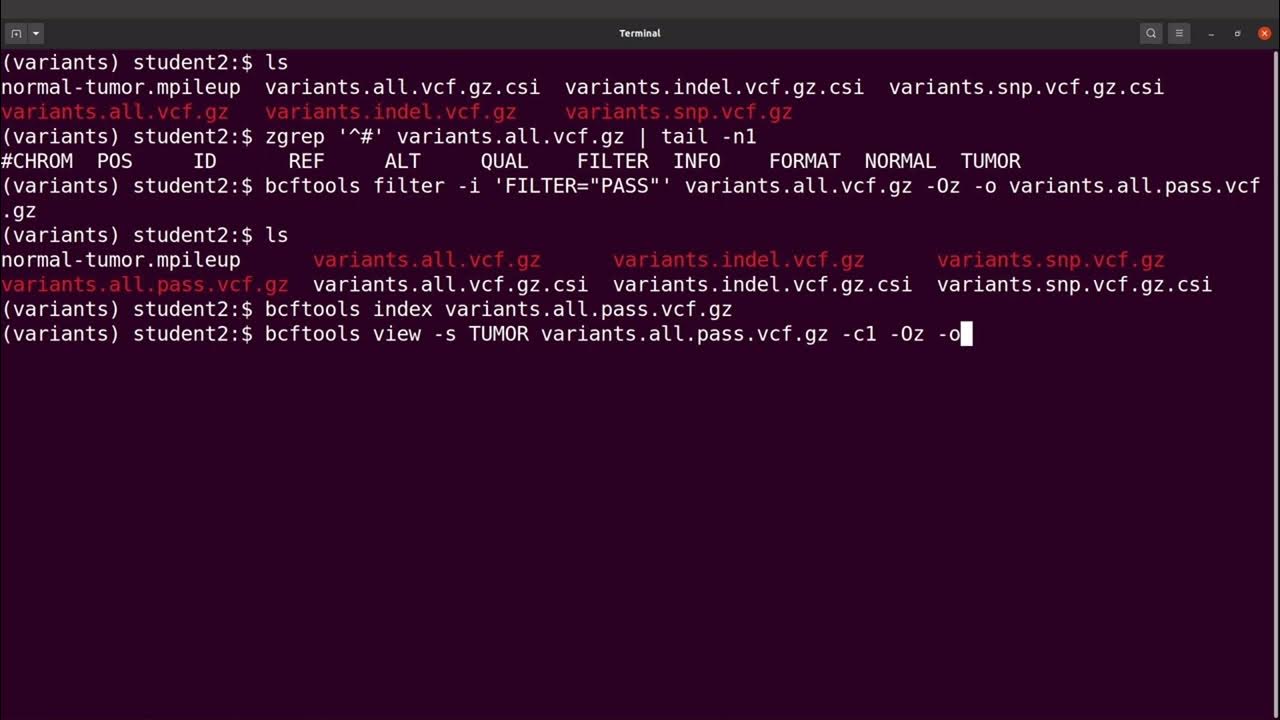
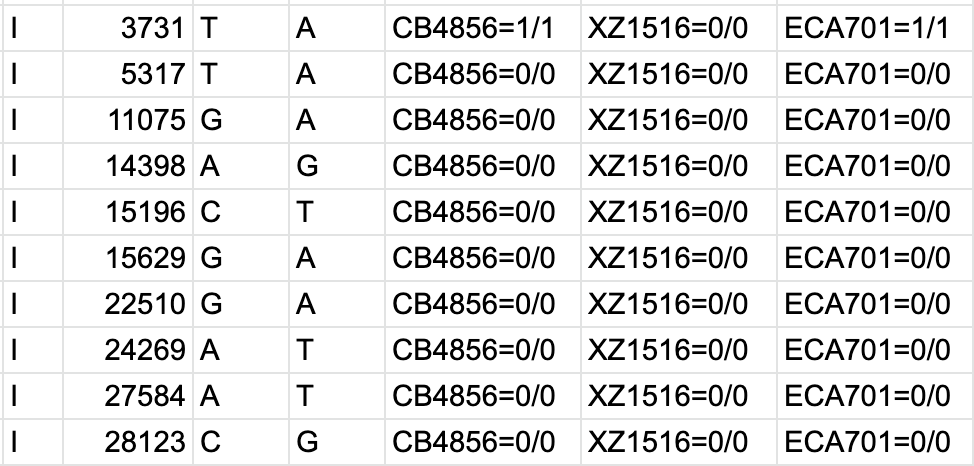
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub

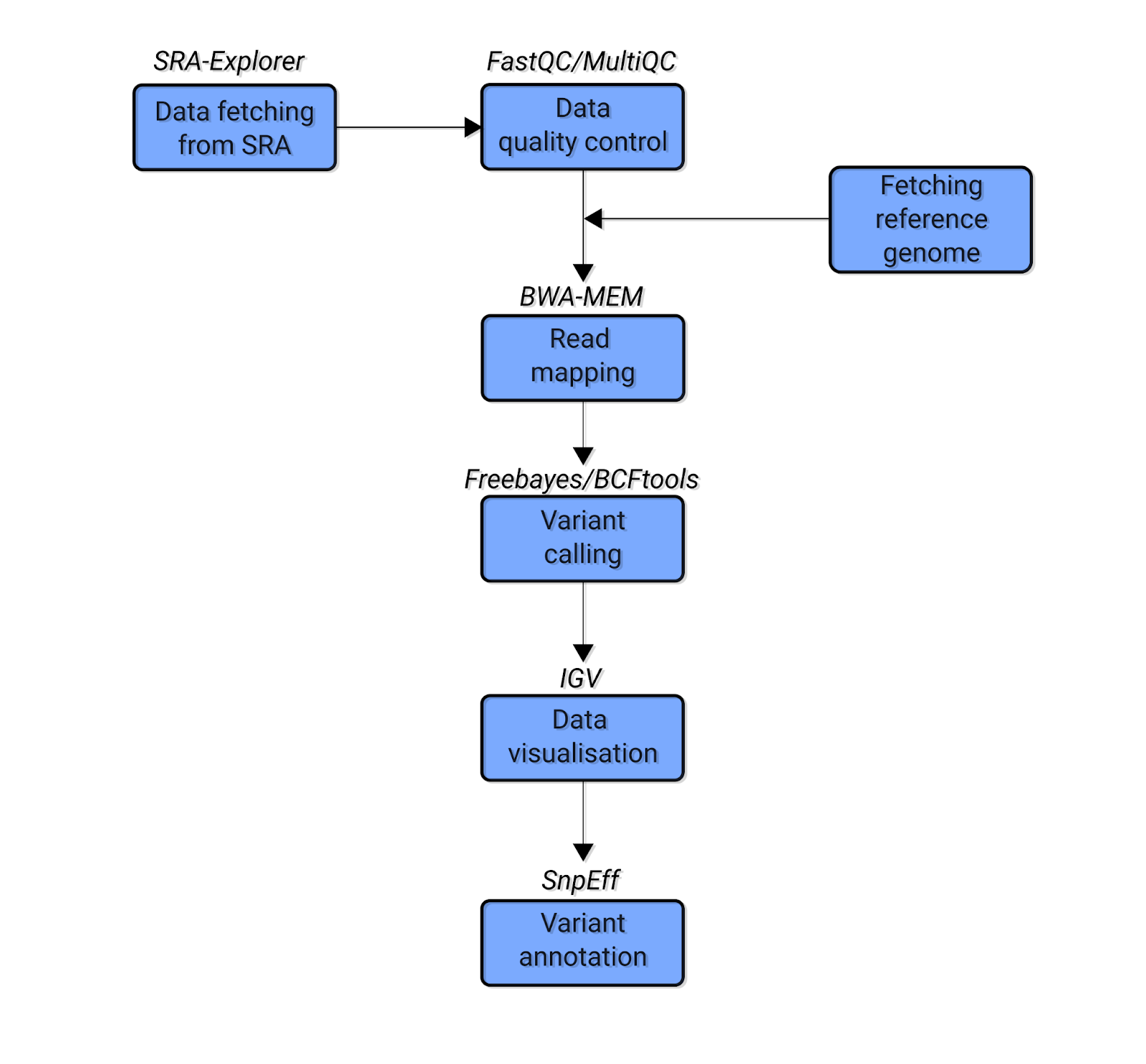
PDF) How to extract and filter SNP data from the genotyping-by- sequencing (GBS) data in vcf format using bcftools