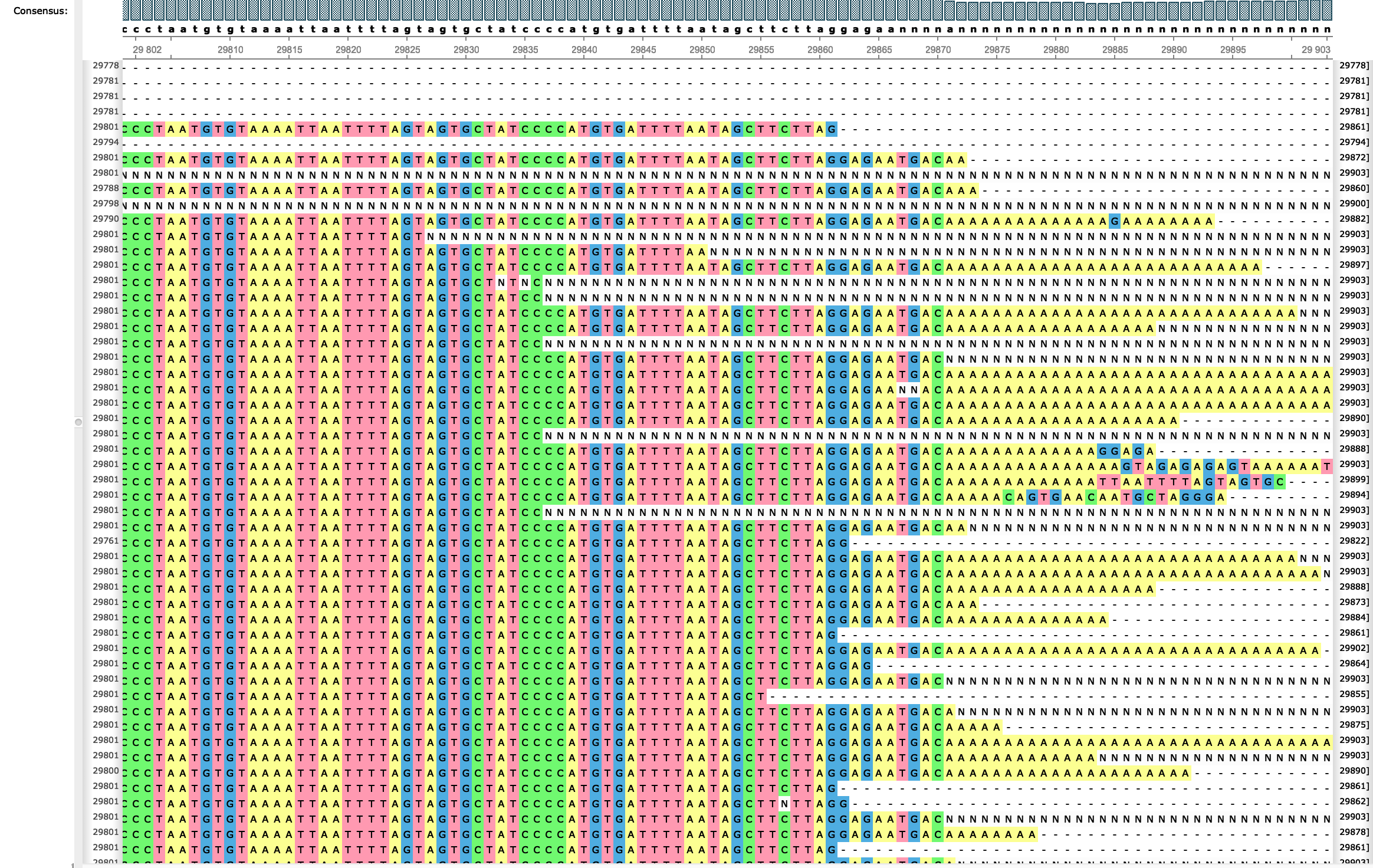
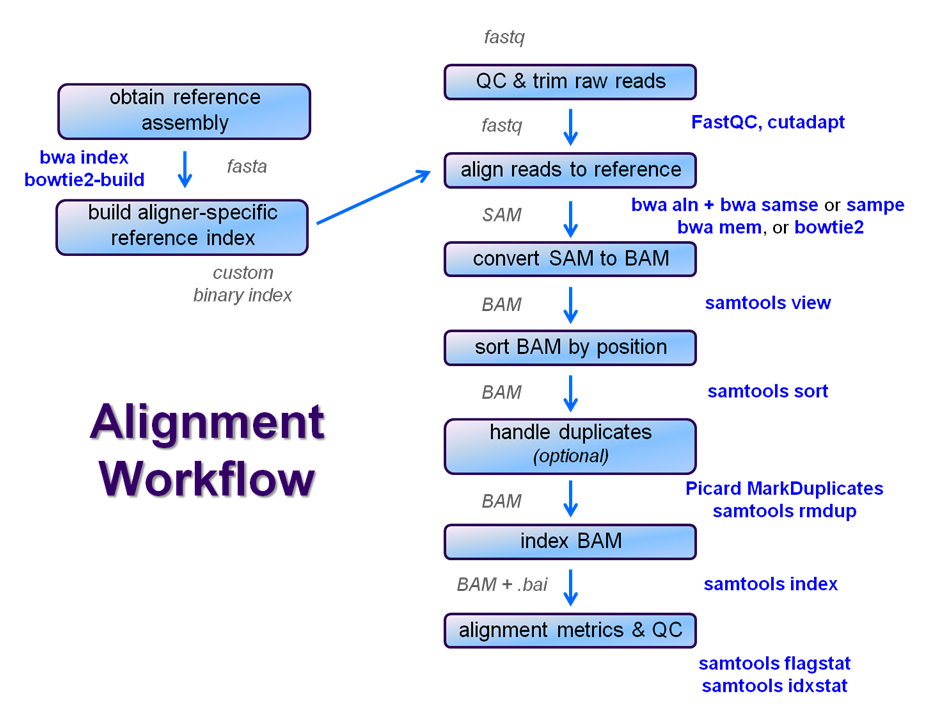
A single test approach for accurate and sensitive detection and taxonomic characterization of Trypanosomes by comprehensive anal

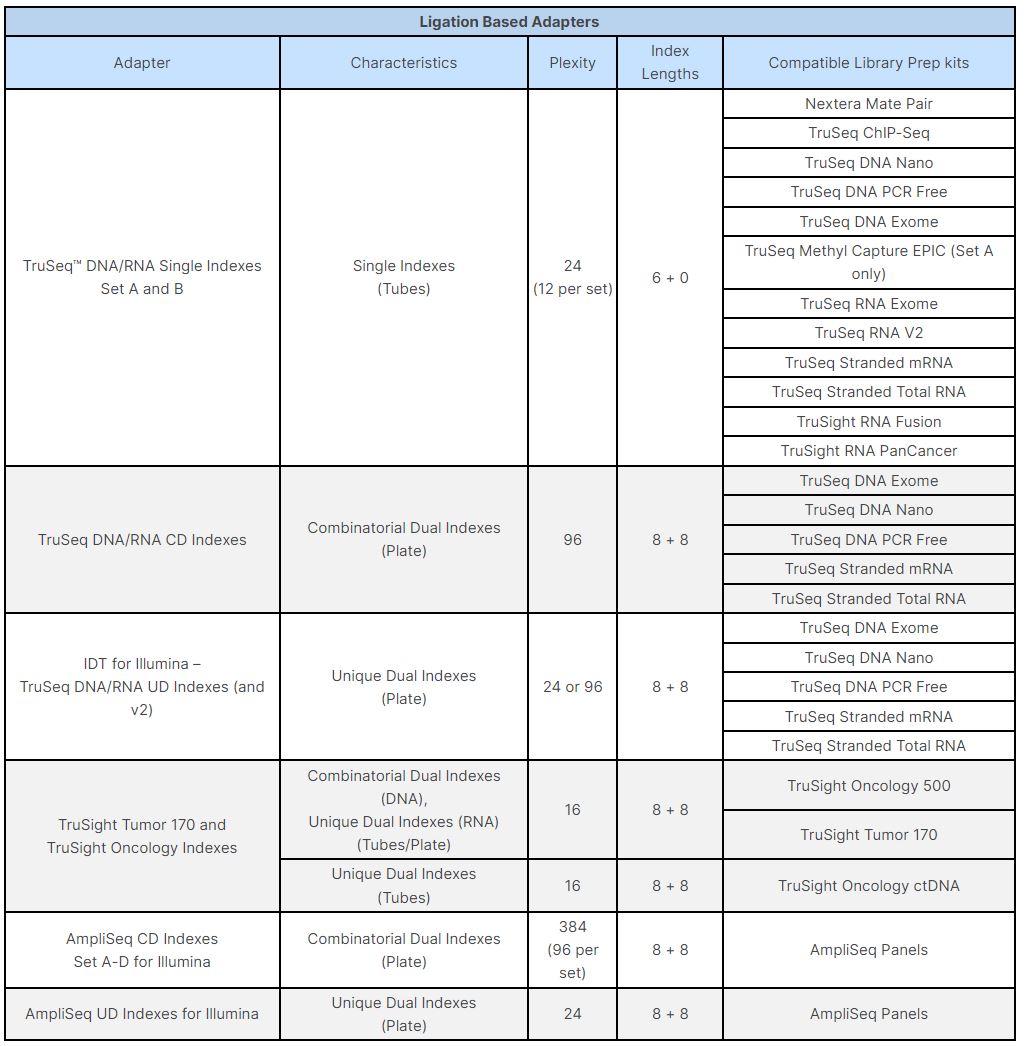
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge
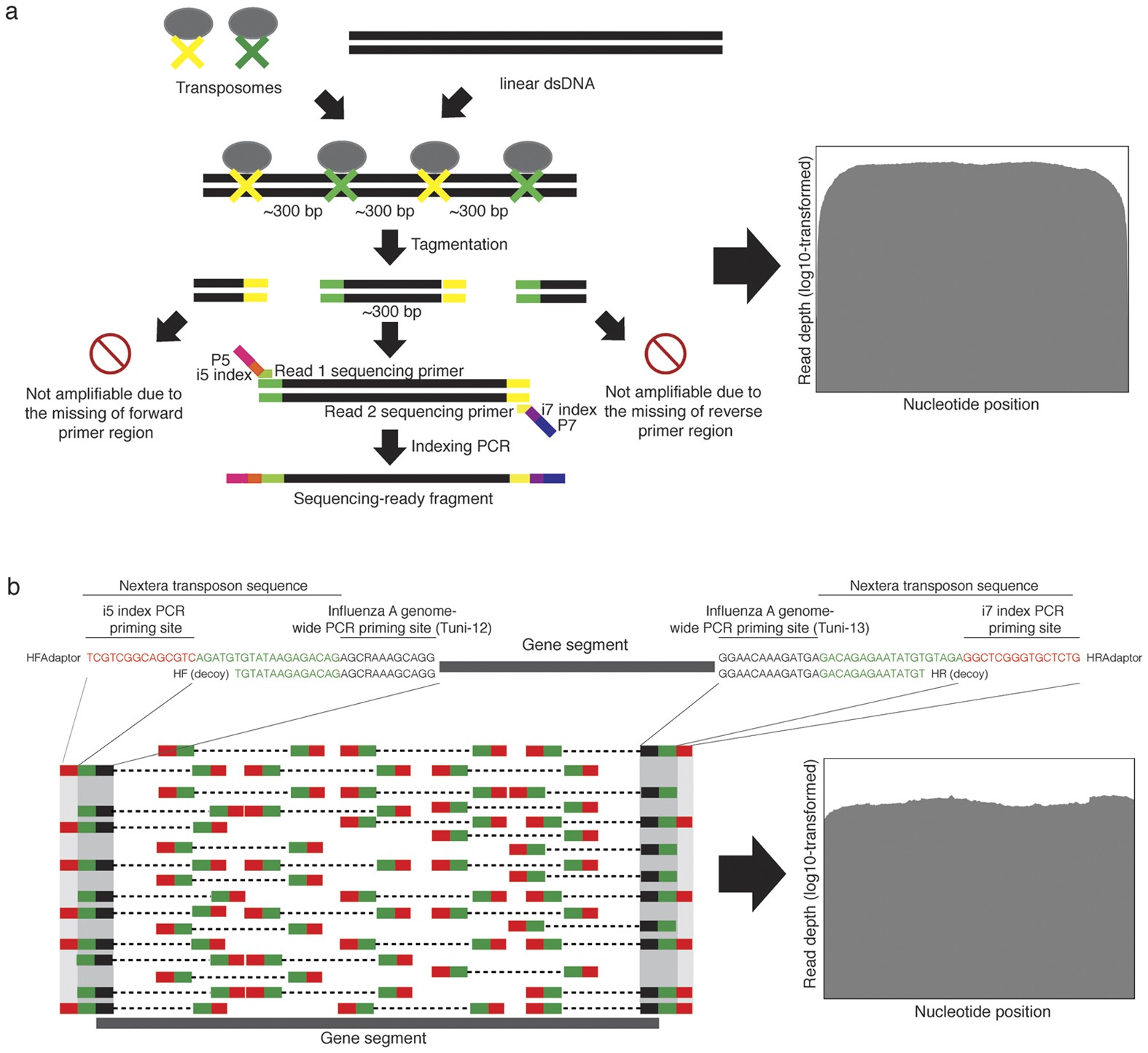
Contamination-controlled high-throughput whole genome sequencing for influenza A viruses using the MiSeq sequencer | Scientific Reports
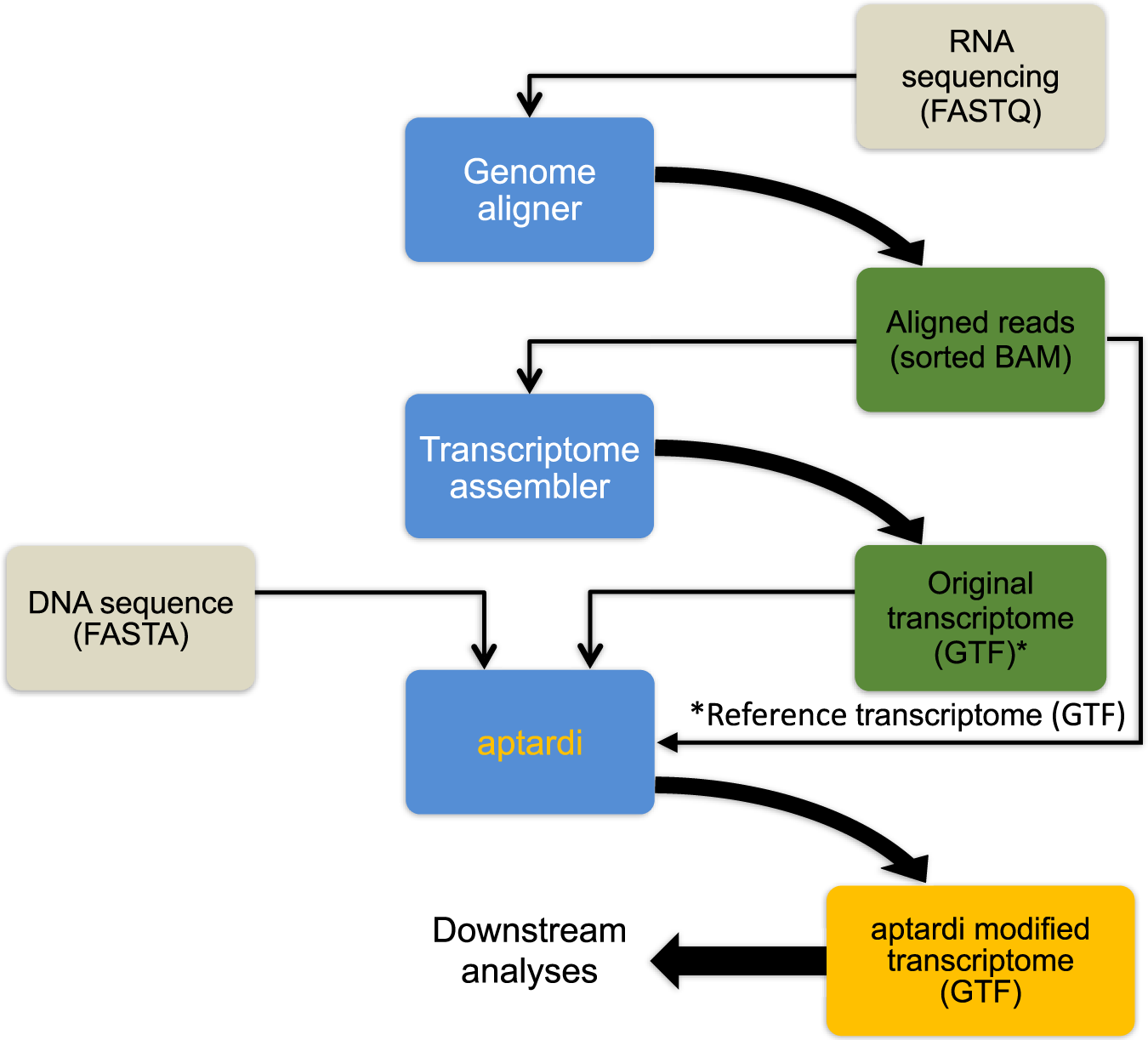
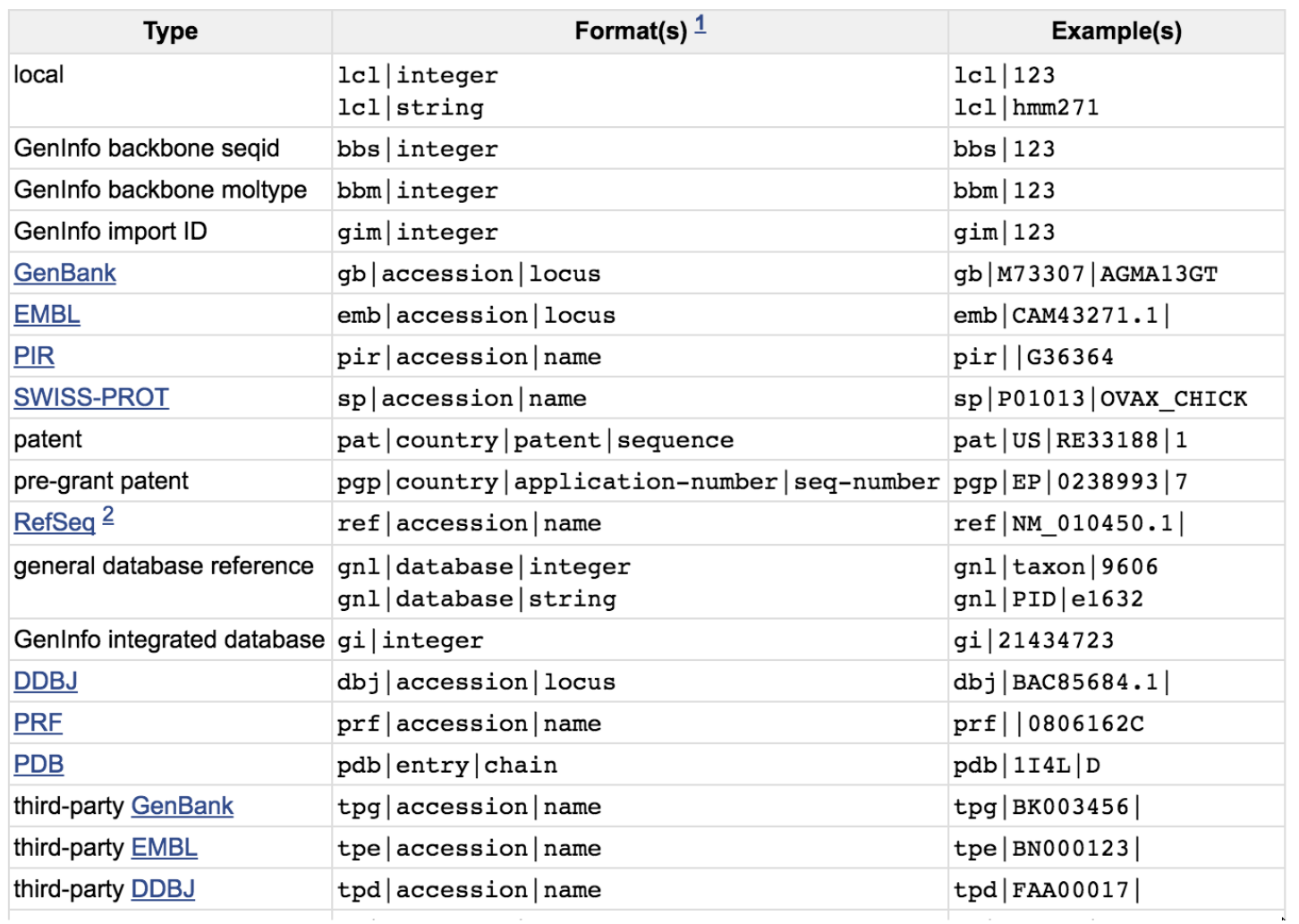
Aptardi predicts polyadenylation sites in sample-specific transcriptomes using high-throughput RNA sequencing and DNA sequence | Nature Communications
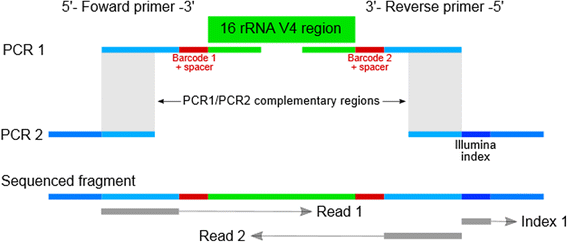
A novel ultra high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform | Microbiome | Full Text

FASTAptamer: A Bioinformatic Toolkit for High-throughput Sequence Analysis of Combinatorial Selections: Molecular Therapy - Nucleic Acids
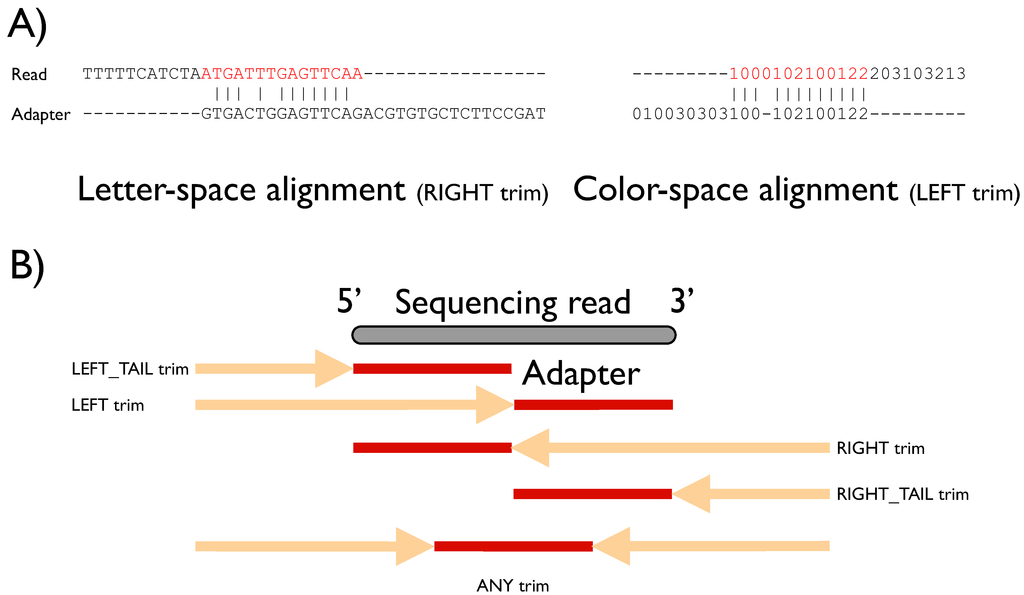
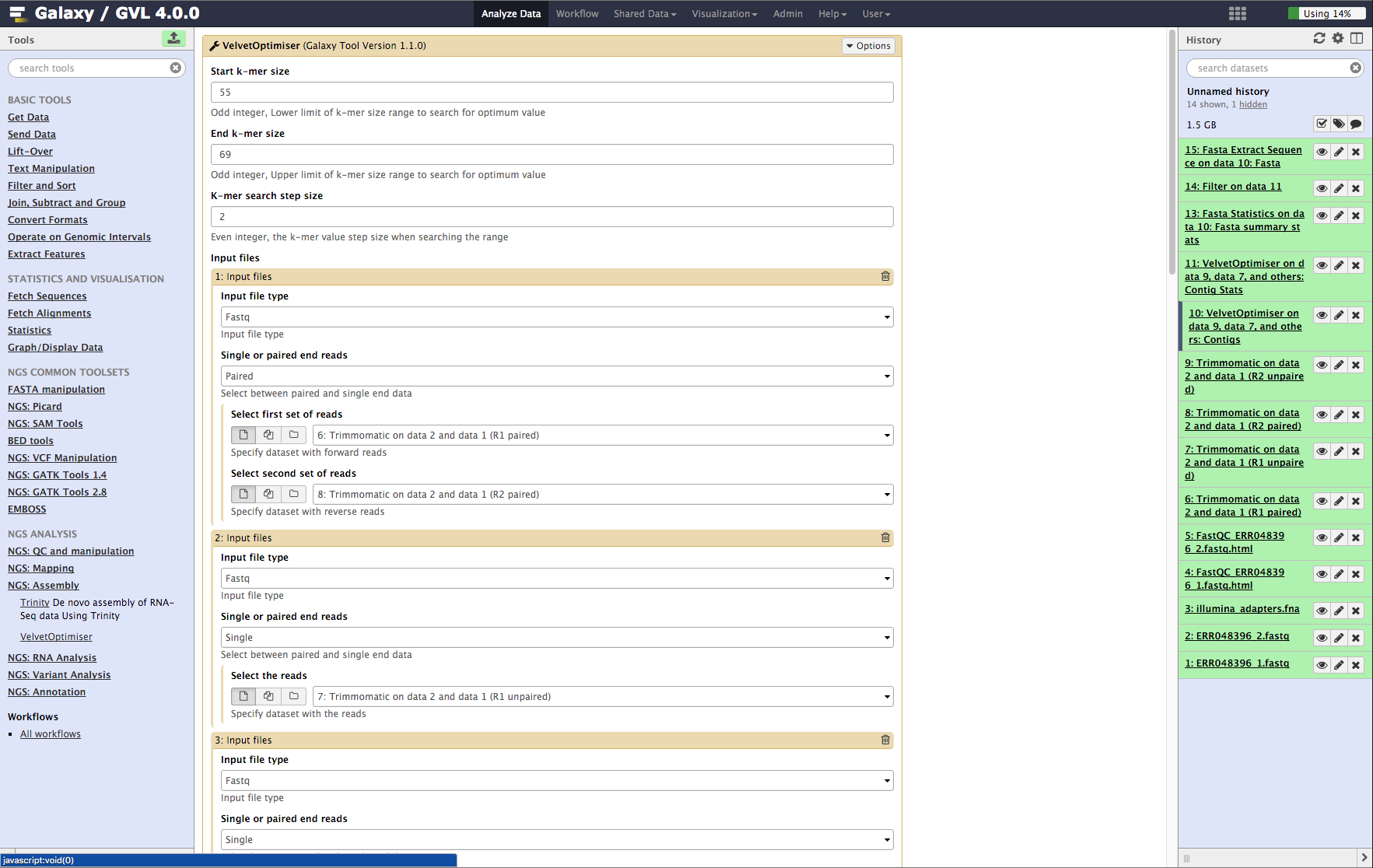
Biology | Free Full-Text | FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms
![SECAPR—a bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments [PeerJ] SECAPR—a bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2018/5175/1/fig-1-full.png)